CommonCrawl on Spark: Reliably Processing 28TB of Data (6300 WARCs, 7 CC Dumps) on a Spark Cluster
Reliably crunch TBs of data with a solid error-tolerant system on Apache Spark!

Being a newbie to the world of big data, I am super-fascinated by what can be done using the CommonCrawl data. Even more with Spark!
To give analogies, CommonCrawl is this all-knowing Oracle from which you can get literally any data present in the visible universe internet and transform it into powerful insights. However, you have to do some heavy lifting to get this. Spark is like Hercules who can wrestle and extract the required information from the Oracle, i.e., CommonCrawl data, effectively at scale.
What We Set Out To Do
In simple words, CommonCrawl is like a snapshot of the internet taken every few months. Hence, you can access almost all the web pages on the internet, and the possibilities of what you can do with that data are endless!
We had an ambitious goal:
- Extract the IPs, hostnames (URLs),
srcattributes present in<script>tags. - Enrich this data with GeoLocation information (coordinates, city, region, etc.) from MaxMind, and run a bunch of analyses on various columns.
- And most importantly, open-source the datasets and the knowledge we gained while doing this exercise!
While we couldn't run all of the planned analyses, we ended up doing a far better job — documenting every step of our process so that anyone can replicate this and continue such endeavors!
This blog post talks about how we designed a fault-tolerant system that can reliably process TBs of data without us having to lose our sleep while staring at the terminal. Here's what I'll be covering:
- a brief note about our Spark setup
- why a fault-tolerant system was needed
- a walk-through of our system (it's simpler than it sounds!)
- the datasets we obtained at the end!
Our Spark Cluster Setup
We went with a dedicated server from Hetzner with the following configuration:
- Intel® Core™ i5-13500 (6P cores, 8E cores)
- 64 GB RAM
- 2 x 512 GB NVMe SSD (Gen4) (Software-RAID 1)
The Spark master and workers were set up as Docker containers with Docker compose orchestrating the containers. I've glossed over so many finer details. Here's a detailed blog talking about our setup:

The server came with a dedicated IPv4 address, everything we did was in Jupyter Lab. So, we port-forwarded the JLab connection over SSH to our laptops.
Why a Fault-Tolerant System?
Here's a diagrammatic overview of our pipeline (without the fault-tolerant or error-handling system):
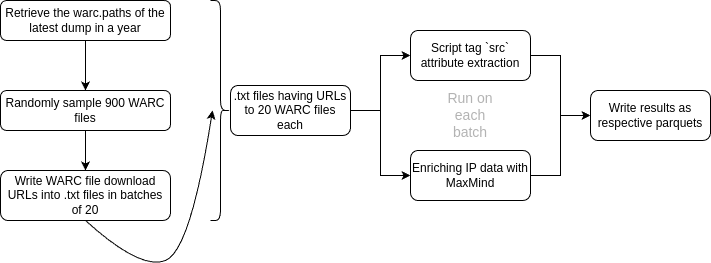
In Pythonic pseudocode, this is how it would look:
year = 2024 # for example
paths: list[str] = download_cc_warc_paths(2024)
# download and sample WARC files
sample_warcs: list[str] = random_sample(paths, 900)
batches: list[list[str]] = batch_samples(sample_warcs, batch_size=20)
# write batches to .txt files
for batch in batches:
write_file(batch, filename="file_i.txt")
# process batches
for batch in batches:
# note that batch only has URLs to WARCs
# and that is why we download to disk first
download_to_disk(batch)
submit_to_spark(script_tag_extr(batch), ipmaxmind(batch))
delete_from_disk(batch)The Problem
There's a problem with this approach. Notice that we are running both the — script extraction and IP geolocation information — jobs in parallel on Spark. What if one (or both) of the jobs fails for whatever reason (we had sufficient error handling in the job scripts themselves, but CommonCrawl used to return 404 for some files)? We'd have very little information to work with to resume the job from where it errored out and don't even get me started on the cleanup we'd have to do for the WARC files and the result files.
But, Suchit, you might ask, why run both jobs in parallel? Why not run one while keeping track of its progress and then run the other? This would result in downloading the batches of WARC files twice, once for each job (which is quite inefficient if you ask me, and we can do better)!
The Solution!
I'll first provide a brief overview of the solution and then go through the implementation.
Overview:
- Have all
.txtfiles (having 20 WARC file URLs each) in a directory, say,warc_splits/and download the WARC files to disk. - Send one batch at a time to both the Spark jobs for processing. Processed data is written to a temporary location, say,
tmp_res/. - Ensure that both jobs have succeeded. A simple way to do so is to have each Spark job script write "success" to a common file, say,
job_status.txtif no error occurs. - If there are 2 "success" in
job_status.txt, then move processed data fromtmp_res/to the permanent location. - Otherwise, we discard the processed results from both jobs and move the
.txtfile (current batch) to another directory, say,unsuccessful/to mark them as, well, unsuccessful. - Remove downloaded WARC files. Each file takes up anywhere between 4-5 GB of disk space. So, we remove them due to disk space constraints.
Implementation Details
I'll now break down, more granularly, how we implemented the entire pipeline.
I won't bore you with some 500 lines of code. Instead, here's the condensed Pythonic pseudocode, whose implementation details can easily be figured out.
Processing warc.paths of each year:
# wp_file -> warc.paths file of a particular year
def process_wp(wp_file: str):
start_time = time.time()
create_dir("warc_splits")
# takes warc.paths file, randomly samples URLs, splits them into batches of 20 each in a .txt file
# we had a simple bash script that downloaded 10 WARC files in parallel with 404 and 5xx error handling
gen_file_splits(wp_file)
for input_txt_file in sorted_files("warc_splits"):
create_dir(data_dir) # data_dir -> directory to store WARC files
# the bash script I talked about earlier
download_warcs(input_txt_file, data_dir)
# after downloading, convert the URLs in the .txt file to
# paths to the respective WARC files on the disk
to_paths(input_txt_file)
# submit the .txt file to Spark for processing (2 jobs).
# successfully processed .txt files are moved to "success/"
submit_job(input_txt_file)
# remove the downloaded WARC files after processing
clear_dir(data_dir)
# move remaining .txt files in "warc_splits" that have
# failed to "unsuccessful/"
move_files("warc_splits", "unsuccessful/")
end_time = time.time()
log_time("times.txt", end_time - start_time)
Submitting to Spark:
def submit_job(input_txt_file: str):
create_dir("tmp_res/")
clear_file(status_file)
# the following two python files have Spark jobs
cmd1 = create_cmd("ipwarc_mmdb_pdudf-errh.py", input_txt_file, "tmp/ipmaxmind_out")
cmd2 = create_cmd("script_extraction-errh.py", input_txt_file, "tmp/script_extraction_out")
# Run jobs as subprocesses
process1 = run_process(cmd1)
process2 = run_process(cmd2)
wait_for(process1, process2)
if both_jobs_successful(status_file):
# move processed data to a permanent location
move_outputs("tmp_res/ipmaxmind_out", f"ipmaxmind_out_{year}")
move_outputs("tmp_res/script_extraction_out", f"script_extraction_out_{year}")
move_file(f"warc_splits/{input_txt_file}", "success/")
log_success(input_txt)
else:
discard_temp_outputs(["tmp_res/ipmaxmind_out", "tmp_res/script_extraction_out"])
log_failure()
File splitting implementation (as is, not pseudocode):
#!/bin/bash
# usage: file_split.sh <input_file> <output_dir> <num_warcs_to_proc>
# Input file containing WARC file paths
input_file=$1
# Output directory for split files
output_dir=$2
mkdir -p "$output_dir"
# Number of warc files to process in the input_file
num_warcs=$3
# WARC year
warc_year=$4
# Check if the input file exists
if [ ! -f "$input_file" ]; then
echo "Input file not found!"
exit 1
fi
# Shuffle the lines in the input file
shuffled_file=$(mktemp)
shuf "$input_file" > "$shuffled_file"
# Number of lines to sample per file
lines_per_file=20
# Total number of output files
total_files=$(($num_warcs / $lines_per_file))
# Create `total_files` number of files with random samples of `lines_per_file` lines each
counter=1
for i in $(seq 1 $total_files); do
output_file="$output_dir/warc_part_$(printf "%03d" "$counter")_$warc_year.txt"
head -n $lines_per_file "$shuffled_file" > "$output_file"
# Remove the selected lines from the shuffled file to avoid duplication
sed -i "1,${lines_per_file}d" "$shuffled_file"
counter=$((counter + 1))
done
# Clean up the temporary shuffled file
rm "$shuffled_file"
echo "File split completed. Files saved in $output_dir"
That is everything that the system does. Super simple, right?
You can find all the code here:
The Processed Dataset
We have completely open-sourced the processed data and it's available on HuggingFace 🤗 for download. We encourage you to tinker with the dataset and can't wait to see what you build with it!

Our dataset has two parts (separated as directories on HF):
- ipmaxmind_out
- script_extraction_out
"ipmaxmind_out" has IPs from response records of the WARC files enriched with information from MaxMind's database.
"script_extraction_out" has IP, server among other columns, and the src attributes of the HTML content in the WARC response records. Visit the HuggingFace repository for more information.
And finally, thank you for reading this blog. I hope you had as much fun reading as I had writing it!




